-Search query
-Search result
Showing 1 - 50 of 528 items for (author: alexandra & t)
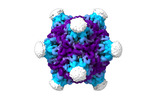
EMDB-19024: 
Structure of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG
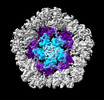
EMDB-19025: 
Structure of the five-fold capsomer of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG
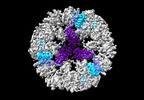
EMDB-19026: 
Structure of the three-fold capsomer of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG
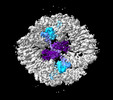
EMDB-19027: 
Structure of the two-fold capsomer of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG

PDB-8rb3: 
Structure of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG

PDB-8rb4: 
Structure of the five-fold capsomer of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG

PDB-8rb5: 
Structure of the three-fold capsomer of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG

PDB-8rb7: 
Structure of the two-fold capsomer of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG

EMDB-16512: 
MiniCoV-ADDomer, a SARS-CoV-2 epitope presenting viral like particle
Method: single particle / : Bufton JC, Capin J, Boruku U, Garzoni F, Schaffitzel C, Berger I

EMDB-16522: 
Structure of ADDoCoV-ADAH11
Method: single particle / : Yadav KNS, Buzas D, Berger-Schaffitzel C, Berger I

PDB-8c9n: 
MiniCoV-ADDomer, a SARS-CoV-2 epitope presenting viral like particle
Method: single particle / : Bufton JC, Capin J, Boruku U, Garzoni F, Schaffitzel C, Berger I

EMDB-16805: 
Cryo-EM structure of PcrV/Fab(30-B8)
Method: single particle / : Yuan B, Simonis A, Marlovits TC

EMDB-16807: 
Cryo-EM structure of PcrV/Fab(11-E5)
Method: single particle / : Yuan B, Simonis A, Marlovits TC

PDB-8cr9: 
Cryo-EM structure of PcrV/Fab(30-B8)
Method: single particle / : Yuan B, Simonis A, Marlovits TC

PDB-8crb: 
Cryo-EM structure of PcrV/Fab(11-E5)
Method: single particle / : Yuan B, Simonis A, Marlovits TC

EMDB-18383: 
Cryo-EM structure of SidH from Legionella pneumophila
Method: single particle / : Sharma R, Weis F, Bhogaraju S

EMDB-18407: 
Cryo-EM structure of SidH from Legionella pneumophila in complex with LubX
Method: single particle / : Sharma R, Adams M, Bhogaraju S

PDB-8qfs: 
Cryo-EM structure of SidH from Legionella pneumophila
Method: single particle / : Sharma R, Weis F, Bhogaraju S

PDB-8qhc: 
Cryo-EM structure of SidH from Legionella pneumophila in complex with LubX
Method: single particle / : Sharma R, Adams M, Bhogaraju S

EMDB-40272: 
BtCoV-422 in complex with neutralizing antibody JC57-11
Method: single particle / : McFadden E, McLellan JS

EMDB-40306: 
BtCoV-422 spike in complex with JC57-11 Fab, global map
Method: single particle / : McFadden E, McLellan JS

EMDB-40310: 
BtCoV-422 spike in complex with JC57-11 Fab, local map
Method: single particle / : McFadden E, McLellan JS

PDB-8sak: 
BtCoV-422 in complex with neutralizing antibody JC57-11
Method: single particle / : McFadden E, McLellan JS

EMDB-17452: 
Single particle cryo-EM structure of the homohexameric 2-oxoglutarate dehydrogenase OdhA from Corynebacterium glutamicum
Method: single particle / : Yang L, Mechaly AM, Bellinzoni M

EMDB-17453: 
Single particle cryo-EM structure of homohexameric 2-oxoglutarate dehydrogenase OdhA from Corynebacterium glutamicum with Coenzyme A bound to the E2o domain
Method: single particle / : Yang L, Mechaly AM, Bellinzoni M

EMDB-17454: 
Single particle cryo-EM structure of homohexameric 2-oxoglutarate dehydrogenase OdhA from Corynebacterium glutamicum in complex with the product succinyl-CoA
Method: single particle / : Yang L, Mechaly AM, Bellinzoni M

EMDB-17455: 
Single particle cryo-EM structure of homohexameric 2-oxoglutarate dehydrogenase OdhA from Corynebacterium glutamicum following reaction with the 2-oxoglutarate analogue succinyl phosphonate
Method: single particle / : Yang L, Mechaly AM, Bellinzoni M

EMDB-17456: 
Single particle cryo-EM structure of the complex between Corynebacterium glutamicum homohexameric 2-oxoglutarate dehydrogenase OdhA and the FHA-protein inhibitor OdhI
Method: single particle / : Yang L, Mechaly AM, Bellinzoni M

PDB-8p5t: 
Single particle cryo-EM structure of the homohexameric 2-oxoglutarate dehydrogenase OdhA from Corynebacterium glutamicum
Method: single particle / : Yang L, Mechaly AM, Bellinzoni M

PDB-8p5u: 
Single particle cryo-EM structure of homohexameric 2-oxoglutarate dehydrogenase OdhA from Corynebacterium glutamicum with Coenzyme A bound to the E2o domain
Method: single particle / : Yang L, Mechaly AM, Bellinzoni M

PDB-8p5v: 
Single particle cryo-EM structure of homohexameric 2-oxoglutarate dehydrogenase OdhA from Corynebacterium glutamicum in complex with the product succinyl-CoA
Method: single particle / : Yang L, Mechaly AM, Bellinzoni M

PDB-8p5w: 
Single particle cryo-EM structure of homohexameric 2-oxoglutarate dehydrogenase OdhA from Corynebacterium glutamicum following reaction with the 2-oxoglutarate analogue succinyl phosphonate
Method: single particle / : Yang L, Mechaly AM, Bellinzoni M

PDB-8p5x: 
Single particle cryo-EM structure of the complex between Corynebacterium glutamicum homohexameric 2-oxoglutarate dehydrogenase OdhA and the FHA-protein inhibitor OdhI
Method: single particle / : Yang L, Mechaly AM, Bellinzoni M

EMDB-27703: 
Structure of RBD directed antibody DH1047 in complex with SARS-CoV-2 spike: Local refinement of RBD-Fab interace
Method: single particle / : May AJ, Manne K, Acharya P

PDB-8dtk: 
Structure of RBD directed antibody DH1047 in complex with SARS-CoV-2 spike: Local refinement of RBD-Fab interace
Method: single particle / : May AJ, Manne K, Acharya P

EMDB-16403: 
Subtomogram averaging of a coronavirus spike (ChAdOx1 19E6) in C3 symmetry
Method: subtomogram averaging / : Ni T, Mendonca L, Zhu Y, Zhang P

EMDB-16404: 
Subtomogram averaging of a coronavirus spike (ChAdOx1 19E6) in C1 symmetry
Method: subtomogram averaging / : Ni T, Mendonca L, Zhu Y, Zhang P

EMDB-16405: 
Subtomogram averaging of a coronavirus spike (ChAdOx1 19E) in C3 symmetry
Method: subtomogram averaging / : Ni T, Mendonca L, Zhu Y, Zhang P

EMDB-16406: 
Subtomogram averaging of a coronavirus spike in situ (ChAdOx1 19E) in C1 symmetry
Method: subtomogram averaging / : Ni T, Mendonca L, Zhu Y, Zhang P

EMDB-16697: 
Subtomogram averaging of a coronavirus spike (ChAdOx1 19E6) in C1 symmetry after 3D classification
Method: subtomogram averaging / : Ni T, Mendonca L, Zhu Y, Zhang P

EMDB-29396: 
Antibody vFP53.02 in complex with HIV-1 envelope trimer BG505 DS-SOSIP
Method: single particle / : Wang S, Kwong PD
EMDB-29836: 
vFP52.02 Fab in complex with BG505 DS-SOSIP Env trimer
Method: single particle / : Gorman J, Kwong PD

EMDB-29880: 
Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 1)
Method: single particle / : Changela A, Gorman J, Kwong PD

EMDB-29881: 
Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 2)
Method: single particle / : Changela A, Gorman J, Kwong PD

EMDB-29882: 
Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 3)
Method: single particle / : Changela A, Gorman J, Kwong PD

EMDB-29905: 
vFP48.02 Fab in complex with BG505 DS-SOSIP Env trimer
Method: single particle / : Gorman J, Kwong PD

PDB-8fr6: 
Antibody vFP53.02 in complex with HIV-1 envelope trimer BG505 DS-SOSIP
Method: single particle / : Wang S, Kwong PD

PDB-8g85: 
vFP52.02 Fab in complex with BG505 DS-SOSIP Env trimer
Method: single particle / : Gorman J, Kwong PD

PDB-8g9w: 
Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 1)
Method: single particle / : Changela A, Gorman J, Kwong PD

PDB-8g9x: 
Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 2)
Method: single particle / : Changela A, Gorman J, Kwong PD
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
